CRISPR/Cas9-mediated gene editing is a powerful technique that allows you to create knock-in/out mutations in any gene and any cell. CRISPR/Cas9 is advantageous over other forms of gene editing, such as TALENs and zinc finger nucleases, because it is simpler to implement and edits at higher efficiency.
Our partners GenScript licenses CRISPR technology developed by the CRISPR pioneering Feng Zhang laboratory. Broad Institute-validated plasmids are a well-tested platform for expressing CRISPR/Cas9, and avoid instability issues in RNA-based from the Broad Institute of MIT and Harvard. Our offerings include the latest CRISPR plasmids and databases platforms.
PRODUCTS
1. CRISPR Plasmid Repository
GenScript maintains a collection of more than 20,000 lentiCRISPRv2 plasmids containing guide RNA (gRNA) sequences pre-validated by the Broad Institute. Plasmids can be searched by gene name, symbol or ID on our gRNA Database.
2. SpCas9 Plasmids
Cas9 endonuclease is the research standard for gene editing. When combined with single guide RNA (sgRNA) sequences, these enzymes create site-specific double strand breaks (DSBs) in the genome.
3. pCas9 Nickase Plasmids
SpCas9 nickase (Cas9n D10A) contains a mutation allowing the endonuclease to create single-strand nicks, as opposed to DSBs. Pairing two opposite facing gRNA sequences with SpCas9 nickase is an efficient method of gene editing that prevents unwanted indels from forming.
4. SaCas9 Plasmids
The Staphylococcus aureus Cas9 orthologue (SaCas9) is the preferred endonuclease for adeno-associated virus (AAV) applications. SaCas9 is approximately 1kb shorter than SpCas9, and offers additional flexibility around AAV packaging constraints. The lower immunogenicity of AAV vectors, makes SaCas9 well-suited for in vivoediting applications and therapeutics.
5. Transcription Activation (SAM) Plasmids
The CRISPR/Cas9 Synergistic Activation Mediator (SAM) system has been engineered to enable transcriptional activation of downstream targets. The SAM system utilizes three different activators, VP64, P65, and HSF1, which are assembled onto a catalytically dead Cas9 (dCas9) complex to drive transcription.
6. Broad Institute Plasmid Collection
Broad Institute Plasmids are generated by the Broad Institute of Harvard and MIT. These plasmids contain a 17bp-1.8kb expressible linker in lieu of a customized sgRNA sequence, which can be modified by your laboratory.
OVERVIEW
SpCas9 Vectors
Cas9 endonucleases derived from the type II CRISPR systems in S. pyogenes (SpCas9) were the first Cas9 enzymes developed for mammalian genome editing. When combined with guide RNA (gRNA) sequences, these enzymes create site-specific double strand breaks (DSBs) in the genome. The CRISPR/Cas9 system accelerated genome editing for its ease of use, specificity, and high efficiency. GenScript is pleased to offer Broad Institute-validated WT SpCas9 constructs for gene editing in mammalian cells. Constructs are available either as all-in-one or dual vector systems, and can be used for non-viral, lenti-viral or adeno-associated virus (AAV) transfection. The lenti-vectors are compatible with 2nd and 3rd generation lentiviral packaging plasmids.
SpCas9 Nickase Vectors
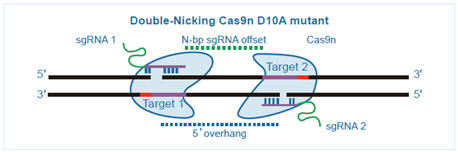 While the CRISPR/Cas9 technology is still more specific when compared to other popular gene editing strategies, off-targeting concerns are still a reality. In an effort to improve specificity, the endonuclease activity of Cas9 was modified. WT Cas9 has two catalytic domains, RuvC and HNH, and mutations to catalytic residues within these domains (specifically, D10A in RuvC and H840A in HNH) cause Cas9 to create single strand nicks as opposed to double strand breaks (Ran et al, 2013). This Cas9 enzyme with nickase activity, or Cas9n, is guided by guide RNAs (gRNA) to opposite sides of the target genomic DNA. Cells will preferentially repair these SSBs by HDR rather than NHEJ. By proceeding through an HDR mechanism, the frequency of unwanted indel mutations from off-target DSBs is minimized. GenScript is pleased to offer Broad-validated nickase vectors for gene editing in mammalian cells types.
While the CRISPR/Cas9 technology is still more specific when compared to other popular gene editing strategies, off-targeting concerns are still a reality. In an effort to improve specificity, the endonuclease activity of Cas9 was modified. WT Cas9 has two catalytic domains, RuvC and HNH, and mutations to catalytic residues within these domains (specifically, D10A in RuvC and H840A in HNH) cause Cas9 to create single strand nicks as opposed to double strand breaks (Ran et al, 2013). This Cas9 enzyme with nickase activity, or Cas9n, is guided by guide RNAs (gRNA) to opposite sides of the target genomic DNA. Cells will preferentially repair these SSBs by HDR rather than NHEJ. By proceeding through an HDR mechanism, the frequency of unwanted indel mutations from off-target DSBs is minimized. GenScript is pleased to offer Broad-validated nickase vectors for gene editing in mammalian cells types.SaCas9 Vectors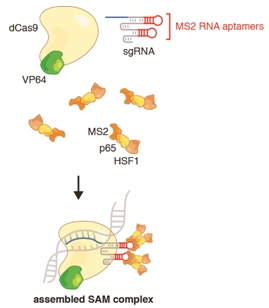

The Cas9 orthologue derived from Staphylococcus aureus, or SaCas9, has similar efficiency to SpCas9; however, SaCas9 is approximately 1kb shorter. The primary advantage of SaCas9 is adeno-associated virus (AAV) packaging: the cargo size of AAV is approximately 4.5kb, and consequently packaging SpCas9 into this vector can be challenging (Ran et al, 2015). The relatively smaller size of SaCas9 makes CRISPR gene editing with AAV vectors possible. Considering the lower immunogenicity of these constructs, SaCas9 is therefore more suited for in vivo editing applications, such as for therapeutics.
Three components -- dCas9-VP64, sgRNA-MS2, and MS2-p-HSF1 – form the assembled SAM complex
Transcription Activation (SAM) Vectors
CRISPR/Cas9 Synergistic Activation Mediator (SAM) is a protein complex engineered to enable robust transcriptional activation of endogenous genes – either a single gene at a time, or up to 10 genes simultaneously in the same cell. SAM takes advantage of the specificity and ease of reprogramming of Cas9 nucleases, which are targeted to a specific locus in the endogenous genome by guide RNA. Through a license with the Broad Institute*, GenScript offers validated SAM gRNA sequences to target any coding region in the human genome, as well as complimentary design of SAM gRNA for any other species. SAM guide RNA sequences are custom-synthesized and cloned into efficient lentiviral vectors, and accompanied by the Cas9-VP64 and MS2-P65-HSF1 components that form the three-part SAM complex.
The SAM complex consists of three components
1. A nucleolytically inactive Cas9-VP64 fusion: dCas9 is used to ensure that no strand breaks are introduced into endogenous genome; VP64 is a transcription activation domain that acts synergistically with p65 and HSF1 to enhance transcription
2. An sgRNA incorporating two MS2 RNA aptamers at the tetraloop and stem-loop: the sgRNA should be designed to target the first 200 bp upstream of the transcription start site in order to target the SAM complex for ideal transcription activation. While sgRNA normally binds to Cas9, the MS2 RNA aptamers are required to allow the third member of the SAM complex to bind to the Cas9-sgRNA complex.
3. The MS2-P65-HSF1 activation helper protein: this contains two transcription activation domains, P65 and HSF1, that synergize with VP64 to robustly activate transcription of downstream coding regions. The MS2 domain allows his helper protein to bind to the sgRNA-dCas9 complex.
CRISPR Vector Specifications
|

